|
|
Journal of Advanced Veterinary Research Volume 9, Issue 2, 2019, Pages 39-44 |
|
|
Molecular Characterization of Biofilm Producing Genes in Salmonellae Isolated from Chicken |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Marwa Ragab Abd El-basit1, Mohamed Wael Abd El-Azeem2, Serageldeen Sultan2, Soad A. Nasef3 |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
1Reference Laboratory for Veterinary Quality Control on Poultry production (RLQP), Animal Health Research Institute, Luxor, Egypt (Corresponding author: drmarwaragab@yahoo.com). 2Department of Microbiology, Faculty of Veterinary Medicine, South Valley University 83523 Qena, Egypt. 3Reference Laboratory for Veterinary Quality Control on Poultry production (RLQP), Animal Health Research Institute, Dokki, Giza 12618, Egypt. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Received: 14 January 2019, Accepted: 10 February 2019 |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Abstract |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Salmonella enterica considered one of the most important food-borne pathogen. Biofilm formation considered one of the main problems related to S. enterica. In this study, biofilm formation, colony morphotype, cellulose and curli production genes of 19 Salmonella isolates were tested. The results showed that 85% of isolates produced strong biofilm and 15% of isolates produced moderate biofilm on polystyrene plate with 1/20 diluted TSB. Different colony morphotypes expressed saw, sbam, and rdar morphotype when cultivated on LB containing Congo red for monitoring cellulose and curli production. All S. enterica strains possess adrA, csgD and gcpA genes using PCR. Thus in this study all Salmonella isolates formed biofilm so they give increased tolerance for antimicrobial agents and disinfectant, which results in difficulty in the treatment of diseases and causing many problems in food industry as it becomes a persistent of source of contamination. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Keywords: Biofilm, curli, cellulose, csgD, adrA, gcpA |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Introduction |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Salmonella is a gram-negative, rod shaped, facultative anaerobic Enterobacteriaceae, it comprises more than 2500 different serovars. Salmonellosis is one of the leading food-borne diseases and a major public health threat worldwide. Localization of bacteria in the environment and in the infected host obtained by the formation of exopolymer matrix known as biofilms (Grantcharova et al., 2010). Microbial biofilm described as an accumulation of cells, mixed species or monospecies, which is attached to a surface and or to each other. Biofilm cells produce proteinaceous substances that protect bacteria from negative environmental condition (Donlan and Costerton, 2002), and become highly resistant to antibiotic (Brown et al., 1988). Salmonella produce biofilms on biotic and abiotic surfaces like plastic, rubber, and stainless steel (Joseph et al., 2001; Solano et al., 2002). Cellulose considered one of the main components of a biofilm, and the other components as curli are responsible for different morphotypes showed by S. enterica on congo red agar. The characteristic RDAR (red, dry and rough) morphotype characterized by cellulose and curli fimbriae production (Chia et al., 2011). This morphotype is closely related to the csgD and adrA genes, which are responsible for the regulation and expression of cellulose, respectively (Fabrega and Vila, 2013). Biofilm bacteria become more resistant to the effect of antimicrobial agent and disinfectant (Joseph et al., 2001; Scher et al., 2005; Wong et al., 2010). Biofilm formed on different food contact surfaces can become a persistent source of contamination leading to foodborne diseases or food spoilage (Shi and Zhu 2009; Vestby et al., 2009). Biofilm formation by S. enterica is an issue of concern in the food industry as Salmonella in biofilms are more protected against antibiotics, disinfectants and environmental stresses than the planktonic cells, which leads to failure in treatment and difficulty of eradication. Thus, this study aimed to detect the ability of Salmonella isolates to form biofilm and the presence of genes responsible for producing biofilm. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Materials and methods |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Strain identification A total of 19 Salmonella strains were isolated from samples that collected from broiler chickens in poultry farms in different localities in Luxor city, Egypt during 2014. Chickens showed suspected signs of salmonellosis, the age of these chickens ranging from 5 to 15 days old. Nine different serotypes were detected (S. kentucky, S. agona, S. bovismorbificans, S. newport, S. vellore, S. sandiego, S. powell, S. nigeria and, S. virchow), the prevalence of these serotypes was 31.6%, 15.8%, 15.8%, 10.5%, 5.26%, 5.26%, 5.26%, 5.26%, 5.26% respectively. Isolation of Salmonella species Salmonella species were isolated and identified biochemically according to ISO-6579:2002 standards. Serological identification of Salmonella Typing of Salmonella isolates was performed according to Kauffman-White scheme (Kauffmann, 1974). Inoculation of Salmonella isolates For inoculation, the strains were transferred from the stock cultures into tryptic soya broth (Difco, MI, USA) and incubated overnight at 37˚C (Stepanovic et al., 2004). The incubated cultures were used for inoculation into different media poured into the wells of plastic microplates (Greiner Bio-One, Germany) for later quantification of biofilm production. Microtitre plate assay The quantification of biofilm formation by Salmonella isolates was assessed using microtitre plate assay as described by Nair et al. (2015). Wells of polystyrene microplate filled with 180 μl of 1/20 diluted tryptic soya broth, 20 μl of cultures of each isolate incubated in TSB dispensed into the wells of polystyrene microplate in triplicate. Negative control wells in triplicate contained 200 μl of broth only. The inoculated plates were incubated aerobically at 37°C for 72 h then the contents of the plates were poured off and the wells thoroughly washed thrice with PBS (pH 7.2). Attached bacterial cells were stained with 200 μl of 0.5% (w/v) crystal violet (Fluka, UK) per well for 10 min. After staining, the plates were washed three times with 300 μl/well of sterile distilled water and allowed air dried, the stained adherent bacterial cells from each well resolved with 250 μL 33% of glacial acetic acid. The optical density (O.D.) of each well was measured at 600 nm using an automated microplate reader (Tecan sunrise, Jencons, UK). The cut-off O.D. (ODc) was defined as three standard deviations above the mean O.D. of the negative control. Thus based on ODc, the Salmonella isolates were classified into four categories: i. Non biofilm producers: O.D. of test isolates ≤ ODc, ii. Weak biofilm producers: O.D. of test isolate ≤ (2×ODc), iii. Moderate biofilm producers: O.D. of test isolate ≤ (4×ODc), iv. Strong biofilm producers: O.D. of test isolate > (4×ODc). Congo red agar plating It was done according to Romling et al. (2003) as bacterial culture plated on LB agar without salt supplemented with 40 µg/ml Congo Red (AppliChem, USA) and 20 µg/ml Coomassie brilliant blue (AppliChem, USA). The colony morphology was determined after incubation at 28°C for 48h. Detection of csgD, adrA and gcpA genes by polymerase chain reaction Nineteen Salmonella isolates were extracted according to QIAamp This work and the used procedures were performed according to the ethical standards of Animal Health Research Institute and Veterinary authorities in Luxor province, Egypt. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Results |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Biofilm formation on polystyrene plates Results demonstrated that all Salmonella isolates produced biofilm when cultivated in the appropriate medium 1/20 diluted tryptic soya broth. Among 19 Salmonella isolates 16 isolates (85%) produced strong biofilm and the other three isolates (15%) produced moderate biofilm as shown in Table 2.
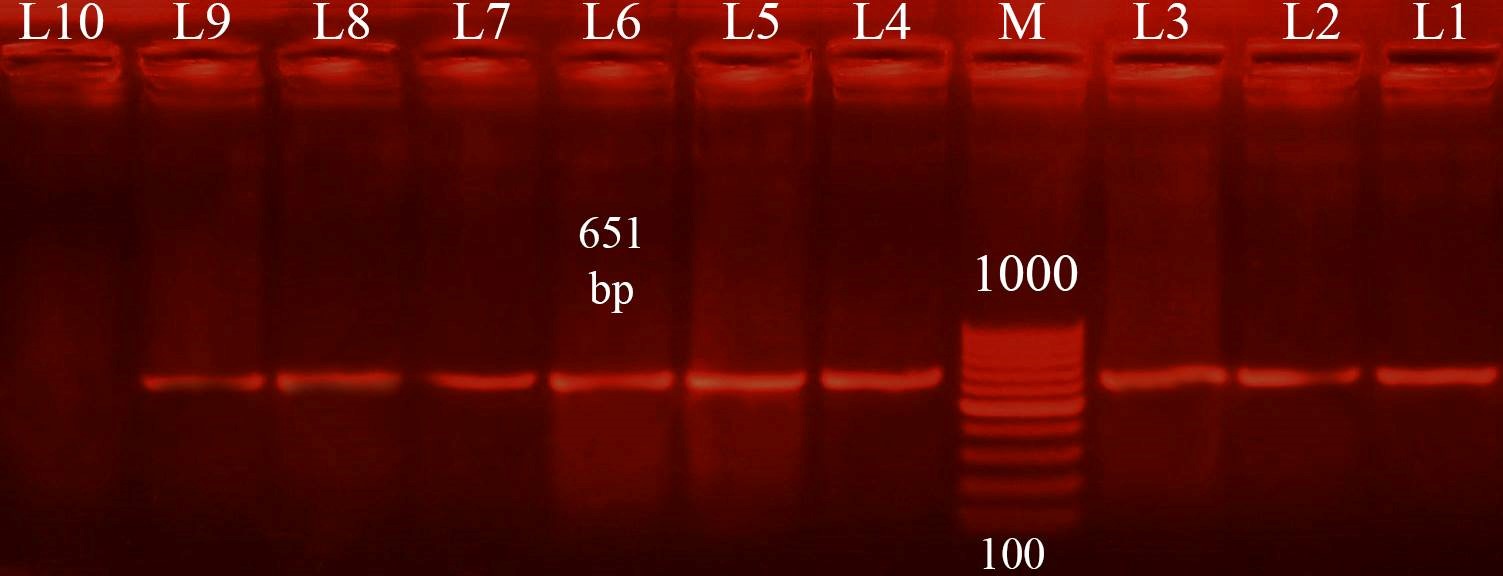
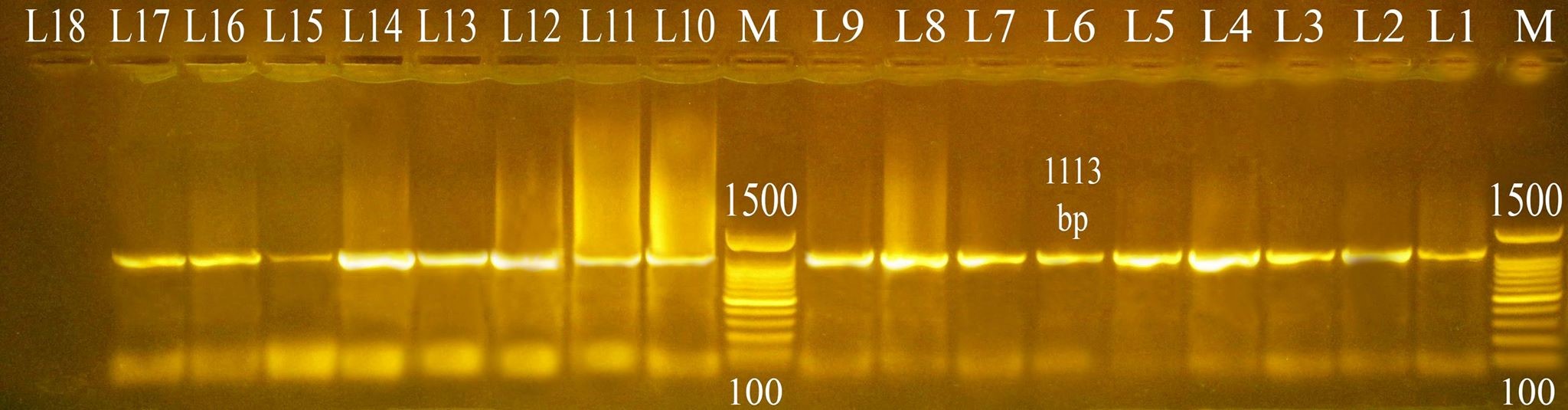
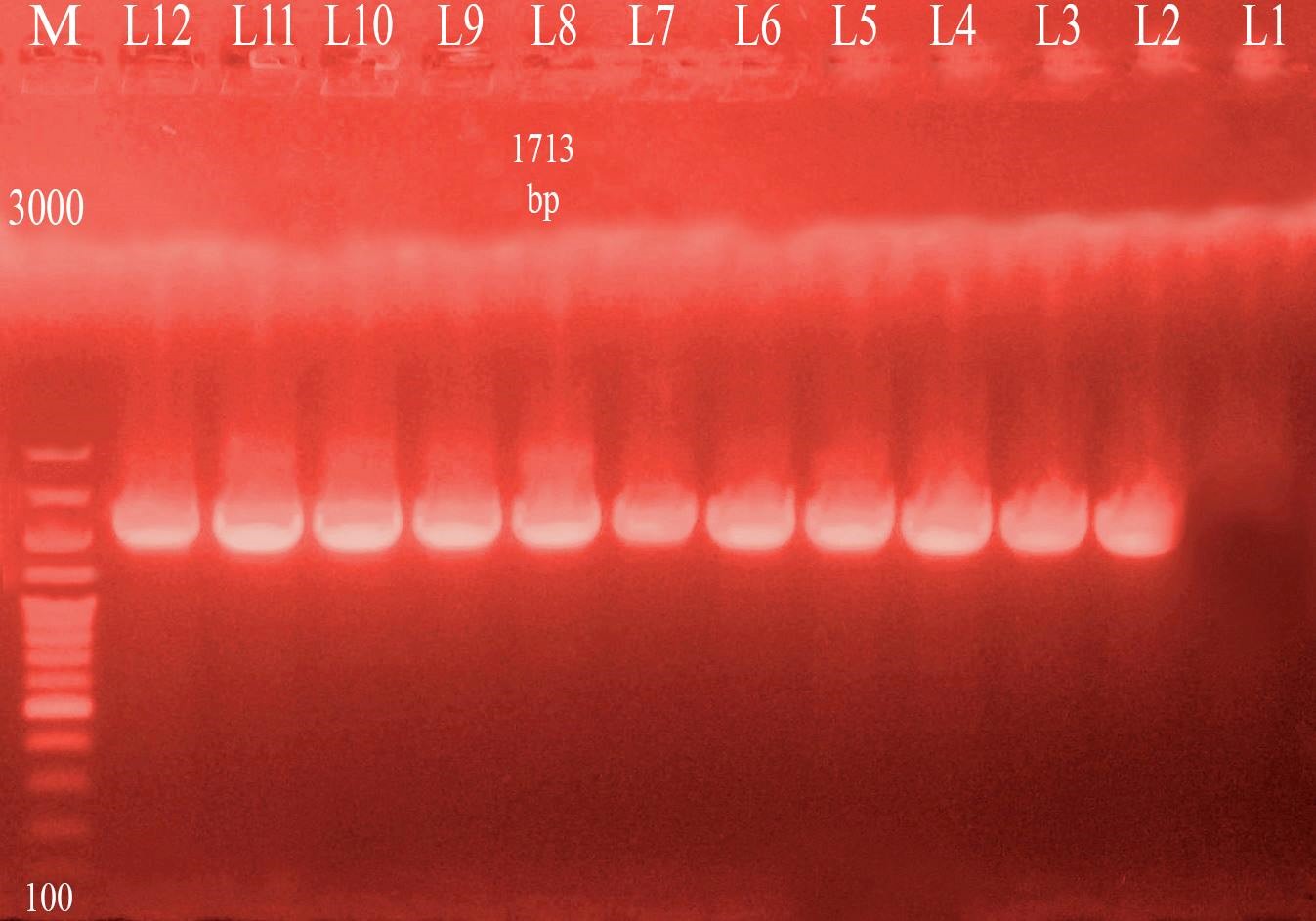
Three different colony morphotypes were detected on Congo red (CR) agar plates (rdar, saw and sbam). rdar (red, dry and rough morphotype) indicating curli and cellulose production, saw (smooth and white morphotype) indicating a lack of both curli and cellulose production finally sbam (smooth brown and mucoid colony) indicating a lack of cellulose synthesis but overproduced capsular polysaccharide. The prevalence of sbam, saw and rdar morphotypes was 52.6%, 31.5%, and 15.7%, respectively. PCR Detection of csgD, adrA, and gcpA All Salmonella isolates analyzed and the results detected that 100 % of isolates were positive for the csgD (Fig. 1), adrA (Fig. 2), and gcpA (Fig. 3) genes. Table 1. Oligonucleotide primers sequences used to detect specific genes
Table 2. Biofilm formation by different Salmonella isolates.
ardar: red dry and rough morphotype; bsaw: smooth and white morphotype; csbam: smooth brown and mucoid colony.
Fig. 2. PCR result for adrA gene (1113 bp) M: 100-1500 bp DNA ladder. Lane 1: positive control sample. Lane 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17: Salmonella isolates positive for detection of adrA gene. Lane 18: negative control sample.
Fig. 3. PCR result for gcpA gene (1713 bp). M: 100-3000 bp DNA ladder.Lane 1:negative control sample. Lane 2,3,4,5,6,7,8,9,10,11. Salmonella isolates positive for detection of gcpA gene. Lane 12: positive control sample. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Discussion |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Salmonella enterica subsp. enterica is one of the main food-borne pathogens, which responsible for 88.715 cases of salmonellosis in 2014 in the European Union (EFSA, 2015), Salmonella spp. considered important pathogenic bacteria, which could be transmitted by food. S. enterica possess recurrent lifestyle that implicated colonization of host with survival in a specific environment. This implies that this pathogen can adapt rapidly from an anaerobic to an aerobic metabolism in order to survive (Encheva et al., 2009). The ability of S. enterica to form biofilms is an issue of concern in food industry, biofilm formation by Salmonella considered important virulence factor. In biofilm cells were more protected against harsh environmental factor, disinfectants, antibiotics and immune system of the host than the planktonic cells (Jensen et al., 2010). Different studies have demonstrated the ability of Salmonella to adhere and form biofilms on a different food contact surfaces, such as metal, plastic and rubber (Joseph et al., 2001; Stepanovic et al., 2004). Biofilm formed in food processing environments had special importance as it became the source of the chronic microbial contamination, which may lead to food spoilage and disease transmission. A biofilm defined as a community of bacterial cells that adhere to a surface and embedded in a self-produced polymeric matrix (Costerton et al., 1999). In the present study, 19 Salmonella isolates investigated for production of biofilm on polystyrene microtiter plate, the results demonstrated that all Salmonella isolates produced biofilm, which in agreement with the reports about Salmonella biofilm on plastic surface obtained by Solomon et al. (2005) and Milanov et al. (2017) as their studies recorded the same results. Lu et al. (2011) found that 39 isolates from 62 S. enterica serovar Pullorum isolates have the ability to produce biofilm, and Malcova et al. (2008) recorded that 80% of Salmonella typhimurium isolates formed biofilm. In this study 85% of isolates were strong biofilm former, while 15% of isolates were moderate biofilm former, while Silva et al. (2014) evaluated the biofilm formation of Salmonella enteritidis on polystyrene and recorded that 25% of isolates were strong biofilm, 35% moderate biofilm, 35% weak biofilm and 10% non-biofilm forming. The differences in biofilm formation could be attributed to strain variations (Chelvam et al., 2014). Nair et al. (2015) found that the majority of isolates of Salmonella formed biofilm (85%), among 40 isolates; 27 isolate were weak biofilm producers and 7 isolates were moderate biofilm producers. Also, other previous studies reported that most of the Salmonella isolates were weak biofilm formers (Diez-Garcia et al., 2012; Ghasemmahdi et al., 2015), while moderate biofilm Salmonella producers were reported by Agarwal et al. (2011) and Naeem et al. (2014). Poultry is considered a major reservoir for S. kentucky (Weill et al., 2006), which then can be transmitted to human from both poultry meat and eggs. Our results indicated that all Salmonella Kentucky isolates form strong biofilm, also Turki et al. (2012) examined 57 Salmonella Kentucky strains isolated from different sources, and the majority of these strains were able to form a biofilm, especially for environmental and animal source isolates. So, the biofilm tends to be a normal and favorable capability in the life of S. kentucky in the environment. S. kentucky was linked with human diseases (81 human cases in 2005), although widespread in the food supply chain, particularly in poultry, in which it has been the most frequent serotype encountered (CDC, 2006; Joerger et al., 2009). Salmonella enterica forms biofilm on biotic and a biotic surface, this Salmonella produces an extracellular matrix with curli and cellulose. Curli are amyloid fibers, which play a role in adhesion to surface, cell aggregation, colonization and biofilm formation (Steenackers et al., 2012). Cellulose, one of the main components of a biofilm which facilitates cell attachment. The characteristic rdar (red, dry, and rough) morphotype, characterized by cellulose and curli fimbriae production, was well studied in S. enterica and allowed it to persist in nutrient-limited environments (Chia et al., 2011). Bacteria produce both curli and cellulose display a red, dry and rough colony morphology (rdar) on Congo red agar plates. Deficiency in curli and cellulose synthesis causes a smooth and white (saw) colony appearance, brown (bdar) colonies obtained by a defect in cellulose synthesis as curli give these brown colony, while the deficiency in curli lead to a pink (pdar) morphotype (Romling, 2005). The result of Congo red in this study indicated that Salmonella spp. showed different morphotypes as three isolates showed rdar, another six showed saw and ten isolates showed sbam morphotype. Similar studies obtained different morphotypes as Malcova et al. (2008) detected different colony morphotypes among 94 strains, 17 strains described saw morphotype, in 10 strains the bdar morphotype was identified and the rdar morphotype was found in 62 strains. Turki et al. (2012) detected the colony morphology for 57 S. kentucky isolates, where five different morphotypes were detected; saw (24), rdar (22), sbam (7), bdar (1) and srad (3). Lu et al. (2011) identified that non biofilm-forming showed nearly white colonies, while biofilm producers showed pink to red colonies. Two morphotype detected in the study of Chia et al. (2011) as all strains display the RDAR morphology except one strain that was a natural and typical BDAR morphotype of this serovar. Lamas et al. (2016a) found that all S. enterica expressed the rdar morphotype in aerobiosis condition and this result disagreed with the present study. S. enterica is in aerobiosis conditions when the bacteria survive outside the host. In these environmental conditions, biofilm formation is an important resource for survival (Fabrega and Vila, 2013). Also Milanov et al. (2017) observed the same result as all isolates of S. tennessee formed the characteristic “rdar” colonies on Congo Red agar. Solano et al. (2002) obtained result near to our result as 93% of Salmonella isolates showed rdar morphotype on CR. Although in this study, all serotypes formed strong and moderate biofilm not all serotypes formed rdar morphotype and mostly of them formed saw and sbam morphotype so the production of curli and cellulose not the only parameter for biofilm formation (Lamas et al., 2016a). Biofilm formation in S. enterica was regulated by different genes. The csgD gene share into biofilm production by its important role in coinciding the expression of several determinants of this process. AdrA gene was essential for biofilm formation by regulating cellulose synthesis through c-di-GMP, which mediates the post transcriptional activation of cellulose biosynthesis (Steenackers et al., 2012). Adhesion of bacteria on surfaces activated the expression of biofilm genes, which are responsible for the fimbriae and cellulose expression in Salmonella. Cellulose and curli were the major constituents of the biofilm matrix; the curli synthesis process is encoded by two operons, csgDEFG and csgBA. CsgA, which is the major curli subunit and CsgB minor subunit encoded by csgBA, csgD is necessary for the transcription csgBA. CsgD also regulated the production of cellulose and requires the expression of adrA gene that activates cellulose synthesis at the post-transcriptional level (Chapman et al., 2002). GcpA (GGDEF domain containing protein A coded by gcpA) protein plays a vital role in inducing biofilm by S. Typhimurium at low nutrient conditions. In this study, we detected the presence of the three genes (adrA, csgD and gcpA) involved in biofilm biosynthesis using PCR and all isolates expressed the three genes. Olivera et al. (2014) agreed with our result as they detected the presence of adrA gene in 112 Salmonella isolates and all of samples positive for adrA gene, which in accordance with our result. Although we have observed a strong association between agfD and adrA genes and the production of biofilm, just a limited proportion of strains were able to produce the characteristic rdar morphology on LB culture medium. The lower percentage of rdar colonies at 28°C (2.3%) could be explained by the observation that thin aggregative fimbriae production in Salmonella spp. was regulated by environmental conditions that play a role on the agfD promoter for triggering the cascade of biofilm production (Romling et al., 1998). In addition, according to Gerstel and Romling (2003), oxygen and pH variation could also interfere on biofilm formation by Salmonella Typhimurium, and directly influence the expression of rdar morphology. Another possibility is that the rdar morphology test is not sensitive enough to detect weak biofilm-producer strains. Also Zeich et al. (2016) agreed with these study assess the presence of the most important to Salmonella biofilm formation genes adrA and csgD in these strains and detected both genes in all examined strains. Seixas et al. (2014) agreed with this study in the presence of adrA and csgD genes 100% of isolates positive for adrA and csgD, and disagreed in detection of gcpA 97.0 % of isolates were positive for gcpA. Bhowmick et al. (2011) detected the genes csgD, adrA and gcpA in all examined isolates, with the exception of SW13 in which adrA and gcpA were negative (Gene expression of gcpA, adrA and csgD was studied by real-time PCR). Lamas et al. (2016b) indicated that among the tested strains, 68.49% (50 of 73) were positive for csgD, 80.82% (59 of 73) were positive for adrA, and 41.10% (30 of 73) were positive for gpcA. The morphotype rdar (red, dry, and rough) was closely related to the csgD and adrA genes, which were responsible for the regulation and expression of cellulose, respectively (Fabrega and Vila, 2013), all Salmonella strains were positive for presence of csgD, adrA and gcpA genes but not all of them expressed rdar morphotype, which indicated that the expression of this morphotype was dependent on certain environmental conditions. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Conclusion |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
The characterization of biofilm formation for different Salmonella isolates revealed that all isolates formed biofilm and different colony morphotype. However, adrA, csgD and gcpA presented in all isolates not all of them expressed the RDAR morphotype. Finally, Biofilm formation allowed the Salmonella to survive within the host and the environment and become persistent source of contamination. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Acknowledgments |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Many thanks to Dr. Waleed Younis, Lecturer of Microbiology, South Valley University, Egypt and Dr. Mohamed Ismail Hassan, Assistant researcher in Animal Health Research Institute, City, Egypt for their support. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Conflict of Interests |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
The authors have declared that no competing interests exist. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
References |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Agarwal, R.K., Singh, S., Bhilegaonkar, K.N., Singh, V.P., 2011. Optimization of microtitre plate assay for the testing of biofilm formation ability in different Salmonella. Int. Food Res. J. 18, 1493-1498. Bhowmick, P.P., Devegowda, D., Ruwandeepika, H.A.D., Fuchs, T.M., Srikumar, S., Karunasagar, I., Karunasagar, I., 2011. gcpA (stm1987) is critical for cellulose production and biofilm formation on polystyrene surface by Salmonella enterica serovar Weltevreden in both high and low nutrient medium. Microbial Pathogenesis 50, 114-122. Brown, M.R., Allison, D.G., Gilber, T.P., 1988. Resistance of bacterial biofilms to antibiotics: a growth-rate related effect?. J. Antimicrob. Chemother. 22, 777–780. CDC., 2006. PHLIS surveillance data, Salmonella annual summary. http://www.cdc.gov/ncidod/dbmd phlisdata/Salmonella.htm. Chapman, M.R., Robinson, L.S., Pinkner, J.S., Roth, R., Heuser, J., Hammer, M., 2002. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295, 851-855. Chelvam, K.K., Chai, L.C., Thong, K.L., 2014. Variations in motility and biofilm formation of Salmonella enterica serovar Typhi. Gut Pathog. 6, 2-10. Chia, T.W.R., McMeekin, T.A., Fegan, N., Dykes, G.A., 2011. Significance of the rdar and bdar morphotypes in the hydrophobicity and attachment to abiotic surfaces of Salmonella Sofia and other poultry-associated Salmonella serovars. Lett. Appl. Microbiol. 53, 581–584. Costerton, J.W., Stewart, P.S., Greenberg, E.P., 1999. Bacterial biofilms: A common cause of persistent infections. Science 284, 1318–1322. Diez-Garcia, M., Capita, R., Alonso-Calleja, C., 2012. Influence of serotype on the growth kinetics and the ability to form biofilms of Salmonella isolates from poultry. Food Microbiol. 31, 173-180. Donlan, R., Costerton, W., 2002. Biofilms: Survival mechanisms of clinically relevant microorganisms. Clinical Microbiology Reviews. 15, 167-193. EFSA (European Food Safety Authority), ECDC (European Centre for Disease Prevention and Control), 2015. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2014. EFSA J. 13, 4329. Encheva, V., Shah, H.N., Gharbia, S.E., 2009. Proteomic analysis of the adaptive response of Salmonella enterica serovar Typhimurium to growth under anaerobic conditions. Microbiol.-SGM 155, 2429–2441. Fabrega, A., Vila, J., 2013. Salmonella enterica serovar typhimurium skills to succeed in the host: virulence and regulation. Clin. Microbiol. Rev. 26, 308–341. Gerstel, U., Romling, U., 2003. The csgD promoter, a control unit for biofilm formation in Salmonella Typhimurium. Research in Microbiology 154, 659 - 667. Ghasemmahdi, H., Tajik, H., Moradi, M., Mardani, K., Modaresi, R., Badali, A., Dilmaghani, M., 2015. Antibiotic resistance pattern and biofilm formation ability of clinically isolates of Salmonella enterica Serotype Typhimurium. Int. J. Enteric Pathog. 3, 27372. Grantcharova, N., Peters, V., Monteiro, C., Zakikhany, K., Romling, U., 2010. Bistable expression of CsgD in biofilm development of Salmonella enteric serovar Typhimurium. J. Bacteriol.192, 456 –466. ISO 6579, 2002. Microbiology of food and animal feeding stuffs horizontal method for the detection of Salmonella species. International Standard., 4th ed. Jensen, P.O., Givskov, M., Bjarnsholt, T., Moser, C., 2010. The immune system vs. Pseudomonas aeruginosa biofilms. FEMS Immunol. Med. Mic. 59, 292–305. Joseph, B., Otta, S.K., Karunasagar, I., 2001. Biofilm formation by Salmonella spp. on food contact surfaces and their sensitivity to sanitizers. Int. J. Food. Microbiol. 64, 367–372. Joerger, R.D., Casey, A.S., 2009. Comparison of genetic and physiological properties of Salmonella enterica isolates from chicken reveals one major difference between serovar Kentucky and other serovars response to acid. Foodborne Pathog. Dis. 6, 503–512. Kauffmann, G., 1974. Kauffmann white scheme. J. Acta. Path. Microbiol. Sci. 61, 385. Lamas, A., Miranda, J.M., Vazquez, B., Cepeda, A., Franco, C.M., 2016a. Biofilm formation, phenotypic production of cellulose and gene expression in Salmonella enterica decrease under anaerobic conditions. International Journal of Food Microbiology 238, 63-67. Lamas, A., Ferandez-No, I.C., Miranda, J.M., Vazquez, B., Ceped, A., Franco, C.M., 2016b. Biofilm Formation and Morphotypes of Salmonella enterica subsp. arizonae differs from those of other Salmonella enterica subspecies in isolates from poultry houses. Journal of Food Protection 79, 1127–1134. Lu, Y., Dong, H., Chen, S., Chen, Y., Peng, D., Liu, X., 2011. Characterization of biofilm formation by Salmonella enterica Serovar Pullorum strains. African Journal of Microbiology Research 5, 2428-2437. Malcova, M., Hradecka, H., Karpiskova, R., Rychlik, I., 2008. Biofilm formation in field strains of serovar Typhimurium: identification of a new colony morphology type and the role of SGI1 in biofilm formation. Veterinary Microbiology 129, 360. Milanov, D., Prunić, B., Ljubojević, D., 2017.Biofilm forming ability of Salmonella enterica serovar Tennessee isolates originating from feed. Vet. Arhiv 87, 691-702. Naeem, A. K., 2014. Detection of swarming and biofilm formation ability of Salmonella Typhimurium isolated from landfills waste. Curr. Res. Microbiol. Biotechnol. 2, 444-449. Nair, A., Rawool, D.B., Doijad, S., Poharkar, K., Mohan, V., Barbuddhe, S.B., Kolhe, R., Kurkure, N.V., Kumar, A., Malik, S.V.S., Balasaravanan, T., 2015. Biofilm formation and genetic diversity of Salmonella isolates recovered from clinical, food, poultry and environmental sources. Infect. Genet. Evol. 36, 424-433. Oliveira de, D.C.V., Fernandes, A., Kaneno, R., Silva, M.G., Araujo, J.P., Silva, N.C.C., Rall, V.L.M., 2014. Ability of Salmonella spp. to produce biofilm is dependent on temperature and surface material. Foodborne Pathog. Dis. 11, 478–483. Romling, U., 2005. Characterization of the rdar morphotype, a multicellular behaviour in Enterobacteriaceae. Cell Mol. Life Sci. 62, 1234-1246. Römlinga, U., Bian, Z., Hammar, M., Sierralta, W.D., Normark, S., 1998. Curli fibers are highly conserved between Salmonella Typhimurium and Escherichia coli with respect to operon structure and regulation. Journal of Bacteriology 180, 722-731. Romling, U., Bokranz, W., Rabsch, W., Zogaj, X., Nimtz, M., Tscha¨pe, H., 2003. Occurrence and regulation of the multicellular morphotypein Salmonella serovars important in human disease. Int. J. Med. Microbiol. 293, 273–285. Romling, U., Sierralta, W.D., Eriksson, K., Normark, S., 1998. Multicellular and aggregative behaviour of Salmonella Typhimurium strains is controlled by mutations in the agfD promoter. Mol. Microbiol. 28, 249–264. Scher, K., Romling, U., Yaron, S., 2005. Effect of heat, acidification, and chlorination on Salmonella enterica serovar Typhimurium cells in a biofilm formed at the air-liquid interface. Appl. Environ. Microbiol. 71, 1163-1168. Seixas, R., Machado, J., Bernardo, F., Vilela, C., Oliveira, M., 2014. Biofilm Formation by Salmonella enterica Serovar 1,4, [5],12:i: Portuguese Isolates: A Phenotypic, Genotypic, and Socio-geographic Analysis. Curr. Microbiol. 68, 670–677. Shi, X., Zhu, X., 2009. Biofilm formation and food safety in food industries. Trends Food Sci. Technol. 20, 407-413. Silva, C. F. da., Gehlen, S.S., Webber, B., Diedrich, L.N., Pilotto, F., Santos, L.R. dos., Tondo, E.C., Nascimento, V.P.do., Rodrigues, L.B., 2014. Biofilm former Salmonella Enteritidis are multiresistant to antibiotics. Acta Scientiae Veterinariae 42, 1229. 27. Solano, C., Garcia, B., Valle, J., Berasain, C., Ghigo, J.M., Gamazo, C., 2002. Genetic analysis of Salmonella Enteritidis biofilm formation: critical role of cellulose. Mol. Microbiol. 43, 793 - 808. Solomon, E.B., Niemira, B.A., Sapers, G.M., Annous, B.A., 2005. Biofilm formation, cellulose production, and curli biosynthesis by Salmonella originating from produce, animal, and clinical sources Journal of Food Protection 68, 906–912 Steenackers, H., Hermans, K., Vanderleyden, J., De Keersmaecker, S.C.J., 2012. Salmonella biofilms: an overview on occurrence, structure, regulation and eradication. Food Res. Int. 45, 502–531. Stepanovic, S., Cirkovic, I.C., Ranin, L., Svabic-Vlahovic, M., 2004. Biofilm formation by Salmonella spp. and Listeria monocytogenes on plastic surface. Lett. Appl. Microbiol. 38, 428–432. Turki, Y., Ouzari, H., Mehri, I., Aissa, R.B., Hassen, A., 2012. Biofilm formation, virulence gene and multi-drug resistance in Salmonella Kentucky isolated in Tunisia. Food Research International 45, 940–946. Vestby, L.K., Moretro, T., Balance, S., Langsrud, S., Nesse, L.L., 2009. Survival potential of wild type cellulose deficient Salmonella from the feed industry. BMC Vet. Res. 5, 43. Weill, F.X., Bertrand, S., Guesnier, F., Baucheron, S., Grimont, P.A.D., 2006. Ciprofloxacin-resistant Salmonella Kentucky in travelers. Emerg. Infect. Dis. 12, 1611–1612. Wong, H., Townsend, K., Fenwick, S., Trengove, R., O’Handley, R., 2010. Comparative susceptibility of planktonic and 3-day-old Salmonella Typhimurium biofilms to disinfectants. J. Appl. Microbiol. 108, 2222-2228. Ziech, R.E., Perin, A.P., Lampugnani, C., Seron, M.J., Viana, C., Soares, V.M., Pereira, J.G., Pinto, J.P.A.N., Bersot, L. dos S., 2016. Biofilm-producing ability and tolerance to industrial sanitizers in Salmonella spp. isolated from Brazilian poultry processing plants. Food Science and Technology 68, 85-90. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|